News
10/30/2024
I’m looking forward to attending NeurIPS 2024 in Vancouver, where we will be presenting preliminary work for two of our most recent projects in workshops this year.
- ProPicker: Promptable Segmentation for Particle Picking in Cryogenic Electron Tomography: Cryogenic Electron Tomography (Cryo-ET) is a crucial imaging technique with the unique ability to image biological macromolecules (e.g. proteins, lipids) in their native cellular environment. We introduce a novel framework for detecting and classifying macromolecules in 3D volumes that greatly accelerates the detection pipeline, and offers rapid adaptation to new and unseen proteins. Our work will be presented as a poster at the Machine Learning in Structural Biology Workshop. This research is the outcome of a fantastic collaboration between our group and Simon Wiedemann and Reinhard Heckel at TUM.
- MediConfusion: Can you trust your AI radiologist? Probing the reliability of multimodal medical foundation models: We present our work on stress-testing the visual reasoning capabilities of medical foundation models at two workshops: AIM-FM: Advancements In Medical Foundation Models and Responsibly Building the Next Generation of Multimodal Foundational Models Workshop
I’m eager to connect with colleagues and engage in discussions about AI for science, healthcare and aspects of reliability!
09/25/2024
Excited to announce MediConfusion, our new medical evaluation benchmark for Multimodal Large Language Models (MLLMs). MediConfusion is a challenging medical Visual Question Answering (VQA) benchmark designed to test the vision capabilities of medical MLLMs. Our findings reveal several critical failures:
- All available models — both open-source and proprietary, even those specifically designed for medical applications — performed below random guessing on MediConfusion.
- State-of-the-art models are easily confused by image pairs that are otherwise visually dissimilar and clearly distinct for medical experts.
- These results raise serious questions about the reliability of current medical MLLMs for healthcare deployment.
We also identified common patterns of model failure, which we hope will guide the development of a new generation of more trustworthy and reliable MLLMs in healthcare. This is just the first step towards a more comprehensive benchmark with more medical subspecialties, categories, and samples yet to come, so stay tuned!
07/15/2024
I’m pleased to share that I’ll be attending ICML 2024 in Vienna, with two main track papers accepted this year, including a Spotlight, as well as some exciting preliminary work.
- [Spotlight] Adapt and Diffuse: Sample-adaptive Reconstruction via Latent Diffusion Models: In this work, we improve the efficiency of diffusion-based inverse problem solvers by adapting the sampling trajectory to the degraded input image. We do this by first estimating how severly degraded the corrupted input is, then we use this degradation severity to find an optimal starting time in the diffusion process. We achieve up to 10x speedup in sampling speed as well as improved reconstruction quality.
- DiracDiffusion: Denoising and Incremental Reconstruction with Assured Data-Consistency: We propose a novel diffusion-based framework for inverse problem solving: we model image corruption as a stochastic degradation process that gradually degrades and noises the clean image. We learn to reverse this corruption process and incrementally add more and more detail back to images. By leveraging early-stopping, we can flexibly trade off perceptual quality (how ‘nice’ the image looks) for better distortion metrics (faithfulness to the observation) or vice versa.
- [Workshop] Serpent: Scalable and Efficient Image Restoration via Multi-scale Structured State Space Models: We explore the potential of Structured State Space Models for image restoration. We introduce a novel architecture that converts the input image into a collection of sequences and leverages state space models as the fundamental computation block. We observe great improvements in model eficiency: reduced compute cost, lower GPU memory requirements and faster inference, and performance on par with state-of-the-art architectures! Our work will be presented at the Next Generation of Sequence Modeling Architectures Workshop.
Looking forward to productive discussions and new connections!
11/20/2023
As NeurIPS 2023 is coming up, I am happy to share that we are going to present some interesting results both in the main track and at various workshops.
- A Data-Free Approach to Mitigate Catastrophic Forgetting in Federated Class Incremental Learning for Vision Tasks: Our work on mitigating catastrophic forgetting in continual learning in a federated setting using a data-free generative approach will be presented as a poster in the main track.
- Adapt and Diffuse: Sample-adaptive Reconstruction via Latent Diffusion Models: Our lab is going to participate in the Deep Learning and Inverse Problems Workshop presenting our recent work on sample-adaptive image reconstruction leveraging latent diffusion models.
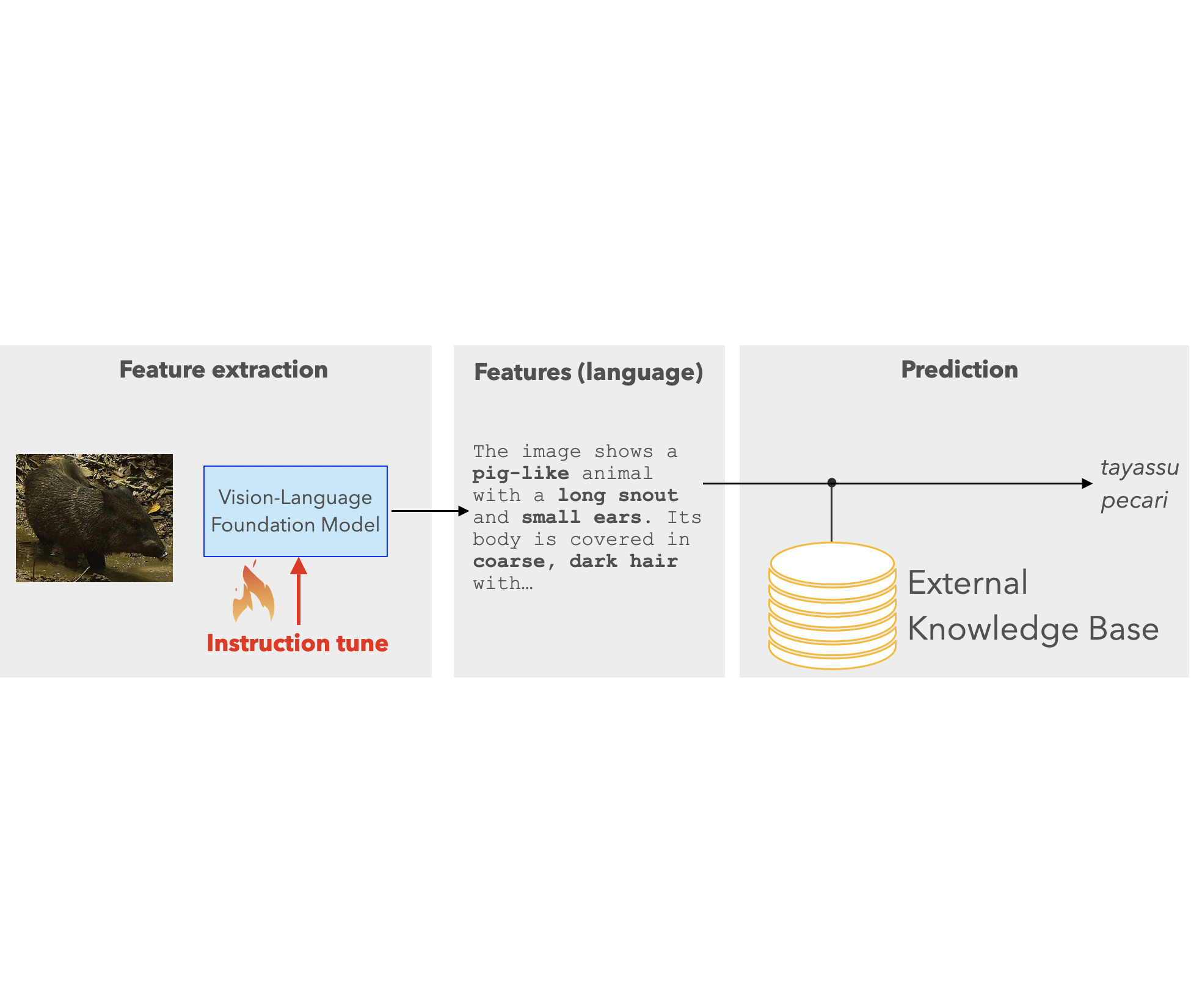
- Multimodal Foundation Models for Zero-shot Animal Species Recognition in Camera Trap Images: My collaboration with Microsoft’s AI for Good lab on zero-shot wildlife recognition using multimodal foundation models will be presented as a poster at the Instruction Workshop.
11/02/2023
After a summer of amazing research collaboration with Microsoft’s AI for Good lab on advancing wildlife conservation efforts, we are eager to announce that a preliminary version of our work has been published on arXiv. We investigate how multimodal foundation models can aid biologists in automatically identifying animal species in camera trap imagery. Our proposed technique, WildMatch, can greatly reduce the cost of image analysis, as it requires no expert-labelled training data. We leverage the rich visual understanding capabilities of pre-trained vision-language foundation models, and adapt them for detailed visual description generation of animals. Then, we find the closest match in an external knowledge base of animal descriptions built from Wikipedia and other publicly available sources. This is still a work in progress with additional results coming soon.
01/15/2023
I am thrilled to share that I’m joining Microsoft’s AI for Good Lab as a research intern for the summer of 2023. Microsoft’s AI for Good initiative is at the forefront of using artificial intelligence to address some of the world’s most pressing global challenges to the environment, humanitarian issues and healthcare. In collaboration with Zhongqi Miao, I will be working on leveraging multimodal foundation models to advance wildlife conservation efforts. I’m excited to learn and make a positive impact with AI for Good researchers!
09/14/2022
Our work HUMUS-Net: a Transformer-convolutional hybrid model for accelerated MRI reconstruction has been accepted for NeurIPS 2022. I am looking forward to sharing our work and interacting with other researchers in the field in person for the first time in a while at NeurIPS.
09/12/2022
I am participating in IPAM’s Computational Microscopy long program at UCLA as a Graduate Visiting Researcher. This program brings together leading experts in the fields of applied mathematics, physics, biology, materials science and engineering in order to encourage debate and collaboration on modern microscopy techniques, such as coherent diffraction imaging, super-resolved fluorescence microscopy (2014 Nobel prize) and a special focus on cryo-electron microscopy (cryo-EM, 2017 Nobel prize). These methods result in high-dimensional, multimodal and extremely noisy data, where extracting useful information, and eventually scientific knowledge is challenging. Deep learning has enormous potential in tackling these challenges that may lead to breakthroughs in materials science, quantum devices and drug discovery.
05/19/2022
I am very excited to share that I will be joining Amazon’s Alexa Perceptual Technologies as an Applied Scientist Intern for the summer under the mentorship of Rajath Kumar. We are going to work on improving wake word verification models through semi-supervised learning techniques and data augmentation. I am looking forward to collaborating with Amazon researchers and learning more about their work.
10/29/2021
Today we hosted the 11th Annual Research Festival at the Ming Hsieh ECE department with close to 100 posters and guided lab tours organized by the Dynamic Imaging Science Center (DISC) showcasing their new, unique high-performance low-field MRI scanner. My poster on MRAugment has won the Best Poster - Honorable Mention award.
09/28/2021
I have been selected as a Ming Hsieh PhD Scholar for 2021-2022 along with Haleh Akrami, Hefei Liu, Rodrigo Lobos, Qinyi Luo and Qiaochu Zhang. We are going to work together to organize professional events and support our PhD community. You can find the slides for my presentation Overcoming the data bottleneck in AI for the sciences presented at the MHI Scholar Finalist Talk Competition below.
07/01/2021
Our paper Data augmentation for deep learning based accelerated MRI reconstruction with limited data has been accepted for short talk at ICML 2021. In this work we propose MRAugment, a data augmentation pipeline for MRI that improves MRI image reconstruction quality when training data is scarce and helps training more robust deep learning models against various forms of distributions shift (different scanner models, anatomies) and hallucinations.
01/18/2021
Our paper 3D phase retrieval at nano-scale via accelerated Wirtinger flow has been accepted at EUSIPCO 2020. High-resolution imaging of small 3D structures is an important problem in biology (protein complexes) and microelectronics (chip manufacturing). Our work introduces a fast and accurate algorithm that can recover the 3D structure of such objects more accurately and from fewer projections than previous techniques.
12/12/2020
Our work Minimax lower bounds for transfer learning with linear and one-hidden layer neural networks has been accepted at NeurIPS 2020 for poster presentation. We investigate how the number of source samples and the distance between datasets impact model generalization on a target dataset. We introduce a novel metric, the so called transfer distance, that quantifies how challenging it is to transfer knowledge from one dataset to another.